What is Functional Data Analysis?
Functional Data Analysis (FDA) is a branch of statistics that deals with data where each observation is a function, curve, or surface rather than a single number or vector. Examples include: - Temperature curves recorded over a day - Growth curves of children over time - Spectrometric measurements across wavelengths - Stock prices throughout trading hours
In FDA, we treat each curve as a single observation and develop methods to analyze collections of such curves.
The fdars Package
fdars (Functional Data Analysis in Rust) provides a comprehensive toolkit for FDA with a high-performance Rust backend. Key features include:
- Fast computation: 10-200x speedups over pure R implementations
- Comprehensive methods: Depth functions, regression, clustering, outlier detection
- Flexible metrics: Multiple distance measures including DTW
- 2D support: Analysis of surfaces in addition to curves
Getting Started
library(fdars)
#>
#> Attaching package: 'fdars'
#> The following objects are masked from 'package:stats':
#>
#> cov, decompose, deriv, median, sd, var
#> The following object is masked from 'package:base':
#>
#> norm
library(ggplot2)
theme_set(theme_minimal())Creating Functional Data
The core data structure is the fdata class. Create
functional data from a matrix where rows are observations (curves) and
columns are evaluation points:
# Generate example data: 20 curves evaluated at 100 points
set.seed(42)
n <- 20
m <- 100
t_grid <- seq(0, 1, length.out = m)
# Create curves: sine waves with random phase and noise
X <- matrix(0, n, m)
for (i in 1:n) {
phase <- runif(1, 0, pi)
X[i, ] <- sin(2 * pi * t_grid + phase) + rnorm(m, sd = 0.1)
}
# Create fdata object
fd <- fdata(X, argvals = t_grid)
fd
#> Functional data object
#> Type: 1D (curve)
#> Number of observations: 20
#> Number of points: 100
#> Range: 0 - 1Adding Identifiers and Metadata
You can attach identifiers and metadata (covariates) to functional data:
# Create metadata with covariates
meta <- data.frame(
group = factor(rep(c("control", "treatment"), each = 10)),
age = sample(20:60, n, replace = TRUE),
response = rnorm(n)
)
# Create fdata with IDs and metadata
fd_meta <- fdata(X, argvals = t_grid,
id = paste0("patient_", 1:n),
metadata = meta)
fd_meta
#> Functional data object
#> Type: 1D (curve)
#> Number of observations: 20
#> Number of points: 100
#> Range: 0 - 1
#> Metadata columns: group, age, response
# Access metadata
fd_meta$id[1:5]
#> [1] "patient_1" "patient_2" "patient_3" "patient_4" "patient_5"
head(fd_meta$metadata)
#> group age response
#> 1 control 54 0.3533851
#> 2 control 43 -0.2975149
#> 3 control 55 0.5553262
#> 4 control 56 -0.3193581
#> 5 control 28 -0.7752047
#> 6 control 38 0.4711363Metadata is preserved when subsetting:
fd_sub <- fd_meta[1:5, ]
fd_sub$id
#> [1] "patient_1" "patient_2" "patient_3" "patient_4" "patient_5"
fd_sub$metadata
#> group age response
#> 1 control 54 0.3533851
#> 2 control 43 -0.2975149
#> 3 control 55 0.5553262
#> 4 control 56 -0.3193581
#> 5 control 28 -0.7752047Key Functionality Overview
Depth Functions
Depth measures how “central” a curve is within a sample. Higher depth indicates a more typical curve:
Distance Metrics
Compute distances between curves using various metrics:
# L2 (Euclidean) distance
dist_l2 <- metric.lp(fd)
# Dynamic Time Warping
dist_dtw <- metric.DTW(fd)Clustering
Group curves into clusters:
# K-means clustering
km <- cluster.kmeans(fd, ncl = 2, seed = 123)
plot(km)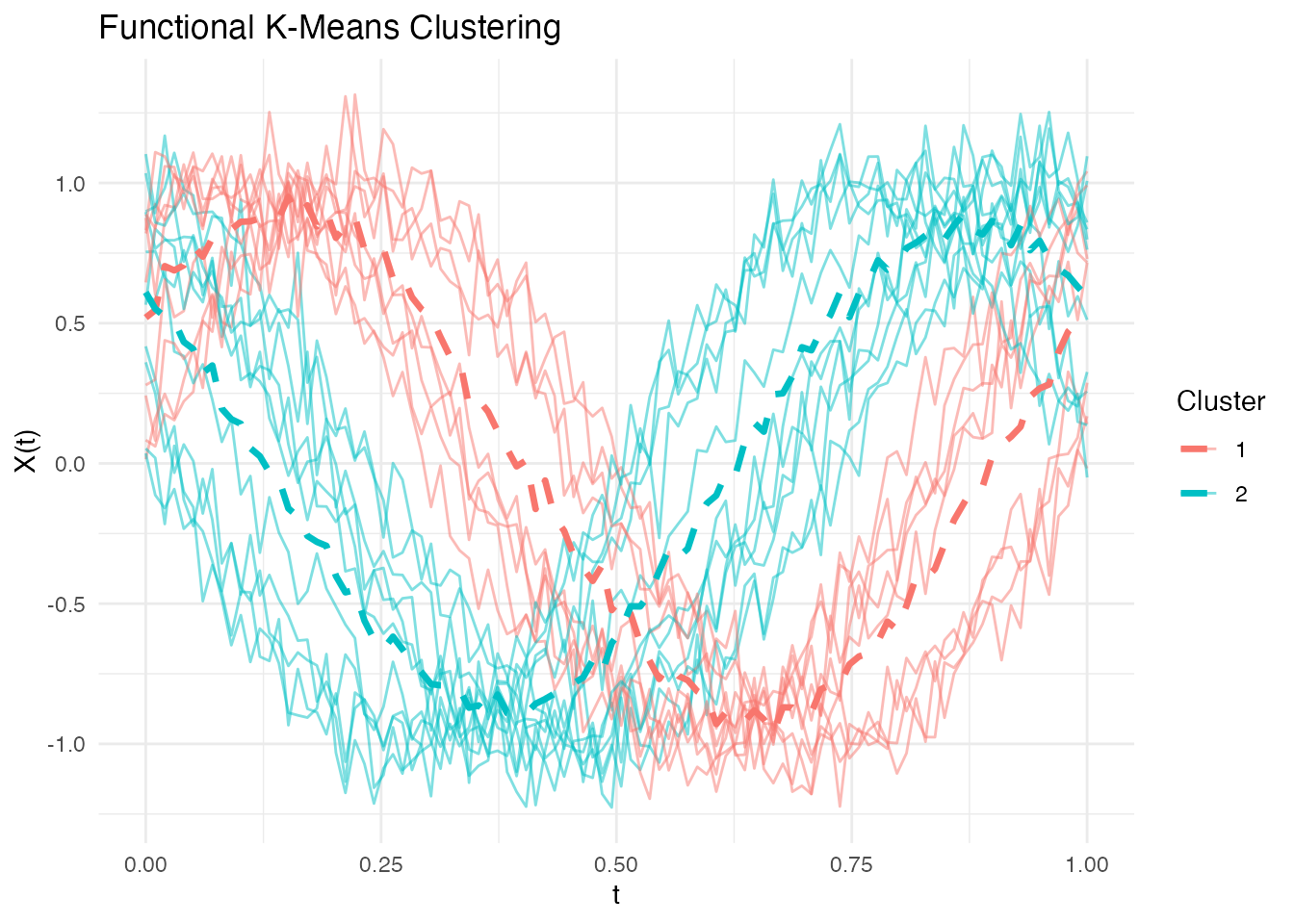
Outlier Detection
Identify atypical curves:
# Add an outlier
X_out <- rbind(X, X[1, ] + 3)
fd_out <- fdata(X_out, argvals = t_grid)
# Detect outliers
out <- outliers.depth.pond(fd_out)
plot(out)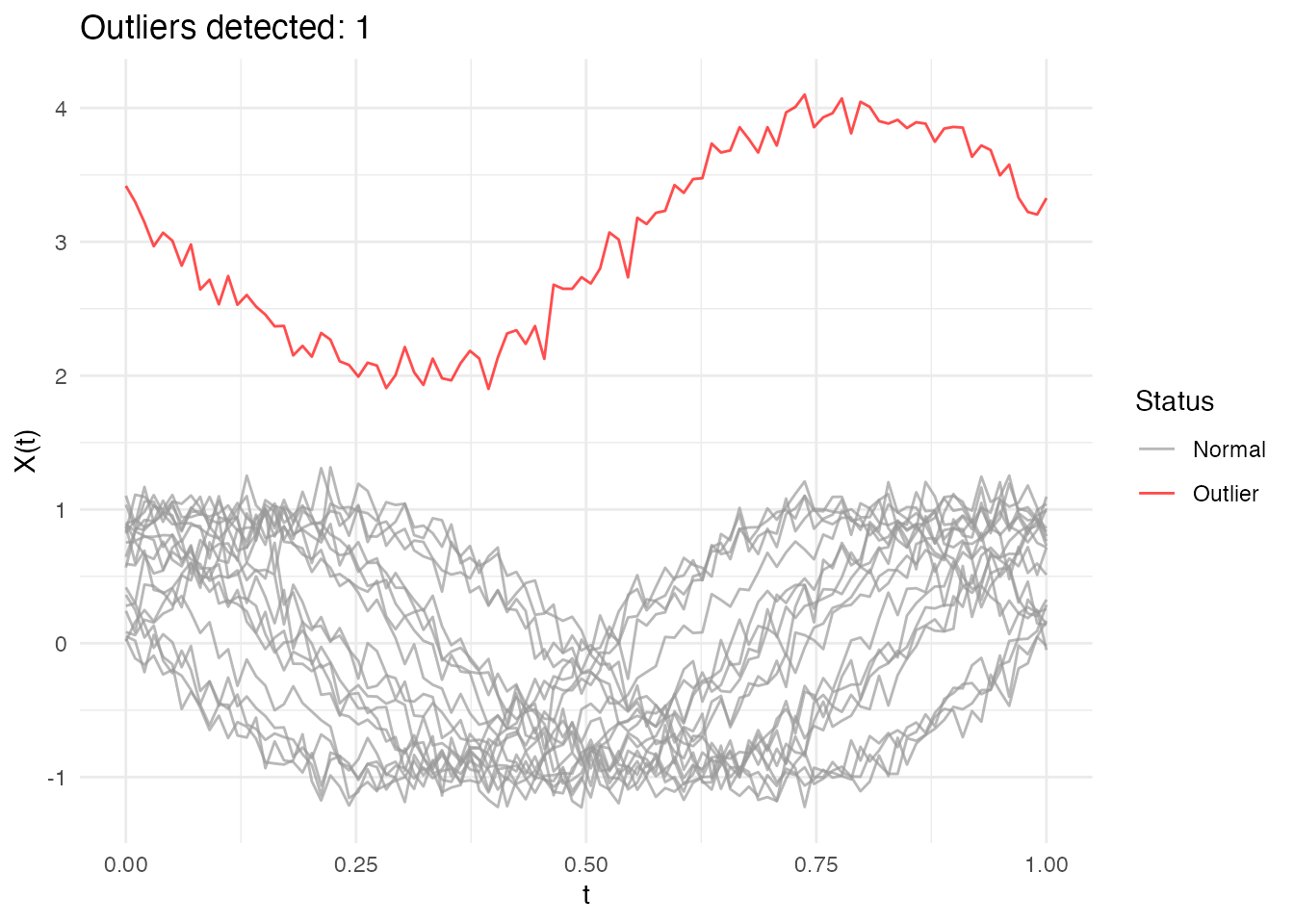
Next Steps
Explore the other vignettes for detailed coverage of specific topics:
- Covariance Functions: Generate Gaussian process samples with various kernels
- Depth Functions: Comprehensive guide to functional depth measures
- Distance Metrics: Distance and semimetric functions
- Regression: Functional regression methods
- Clustering: Functional k-means and optimal k selection
- Outlier Detection: Methods for identifying atypical curves
Performance
The Rust backend provides significant speedups for computationally intensive operations. For example, computing depth for 1000 curves:
# Generate large dataset
X_large <- matrix(rnorm(1000 * 200), 1000, 200)
fd_large <- fdata(X_large)
# Depth computation is fast even for large datasets
system.time(depth(fd_large, method = "FM"))
#> user system elapsed
#> 0.045 0.000 0.045References
- Ramsay, J.O. and Silverman, B.W. (2005). Functional Data Analysis. Springer.
- Ferraty, F. and Vieu, P. (2006). Nonparametric Functional Data Analysis. Springer.
- Febrero-Bande, M. and Oviedo de la Fuente, M. (2012). Statistical Computing in Functional Data Analysis: The R Package fda.usc. Journal of Statistical Software, 51(4), 1-28.